 |
ATAC-seq
|
 |
PUBLISHED ON: JANUARY 02, 2017 | REV C
SUPPORTED IFCS: Open App IFC
NUMBER OF IFC RUNS: 17
Overview
Cell-to-cell variation is a universal feature of life that impacts a wide range of biological phenomena, from developmental plasticity to tumor heterogeneity. While recent advances have improved our ability to document cellular phenotypic variation the fundamental mechanisms that generate variability from identical DNA sequences remain elusive. Here we reveal the landscape and principles of cellular DNA regulatory variation by developing a robust method for mapping the accessible genome of individual cells via assay for transposase-accessible chromatin using sequencing (ATAC-seq). Single-cell ATAC-seq (scATAC-seq) maps from hundreds of single cells in aggregate closely resemble accessibility profiles from tens of millions of cells and provide insights into cell-to-cell variation. Accessibility variance is systematically associated with specific trans-factors and cis-elements, and we discover combinations of trans-factors associated with either induction or suppression of cell-to-cell variability. We further identify sets of trans-factors associated with cell-type specific accessibility variance across 8 cell types. Targeted perturbations of cell cycle or transcription factor signaling evoke stimulus-specific changes in this observed variability. The pattern of accessibility variation in cis across the genome recapitulates chromosome topological domains de novo, linking single-cell accessibility variation to three-dimensional genome organization. All together, single-cell analysis of DNA accessibility provides new insight into cellular variation of the regulome. This script has been updated to be compatible with all versions of the C1™ Open App™ IFCs, including the redesigned medium 96-cell IFC.
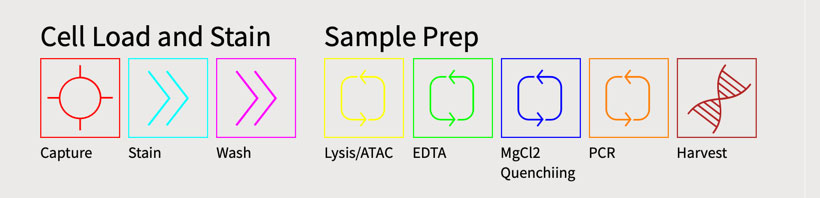
Protocol: ATAC-seq | Duration (H:M): 2:00
Tested Primary Cells or Cell Lines
Cell Name |
Cell Type |
Source |
|---|---|---|
| K562 | Leukemia | Cell line |
| GM12878 | B-Lymphocyte | Cell line |
| HL-60 | Promyeloblast | Cell line |
| EML | Basophil | Cell line |
| TF-1 | Erthroblast | Cell line |
| H1 ESC | Stem cell | Cell line |
| BJ | Fibroblast | Cell line |
| mESC | Stem cell | Cell line |
Performance
Data from single cells recapitulate several characteristics of bulk ATAC-seq data, including fragment size periodicity corresponding to integer multiples of nucleosomes, and a strong enrichment of fragments within regions of accessible chromatin. Microfluidic chambers generating low library diversity or poor measures of accessibility, which correlate with empty chambers or dead cells, were excluded from further analysis. Chambers passing filter yielded an average of 7.3x104 fragments mapping to the nuclear genome.
Resources
Publications or Articles
Additional documents
Unless explicitly and expressly stated otherwise, all products are provided for Research Use Only, not for use in diagnostic procedures. Find more information here.
