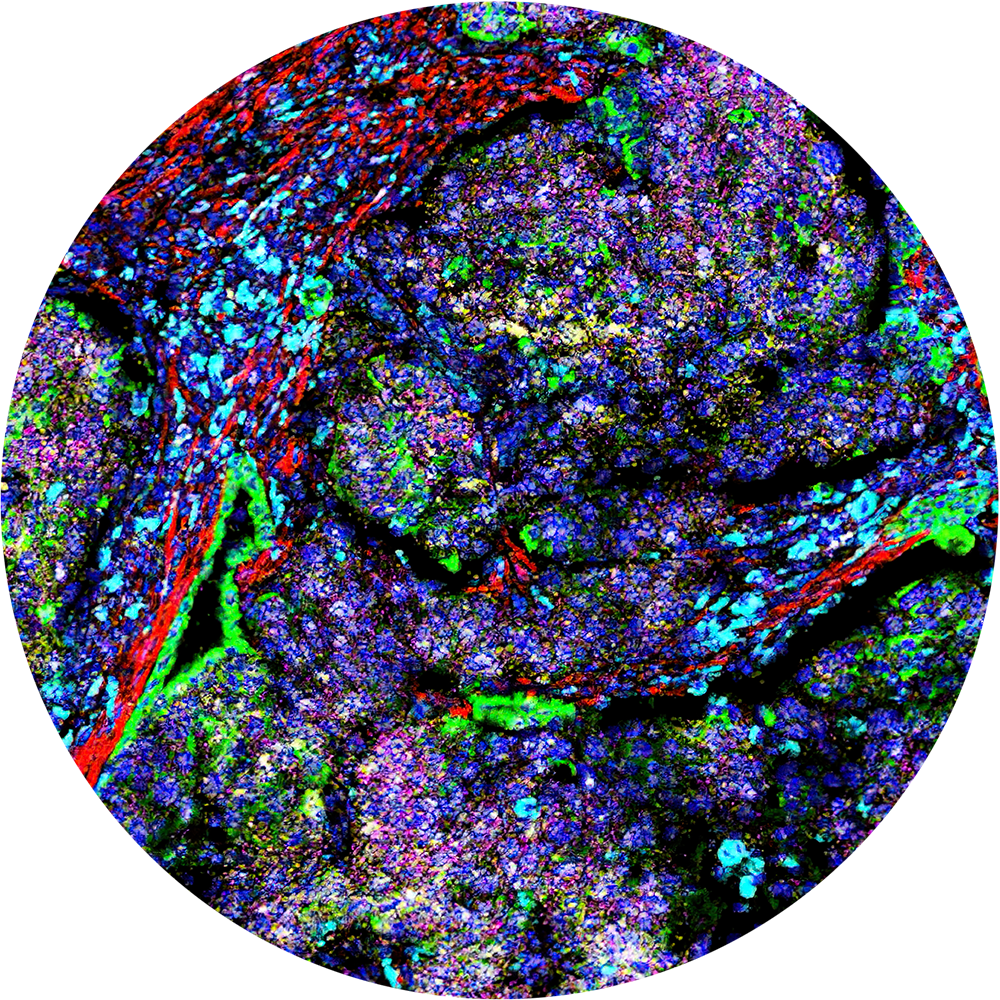
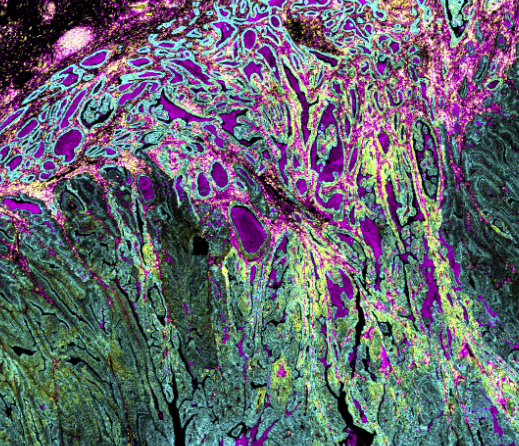
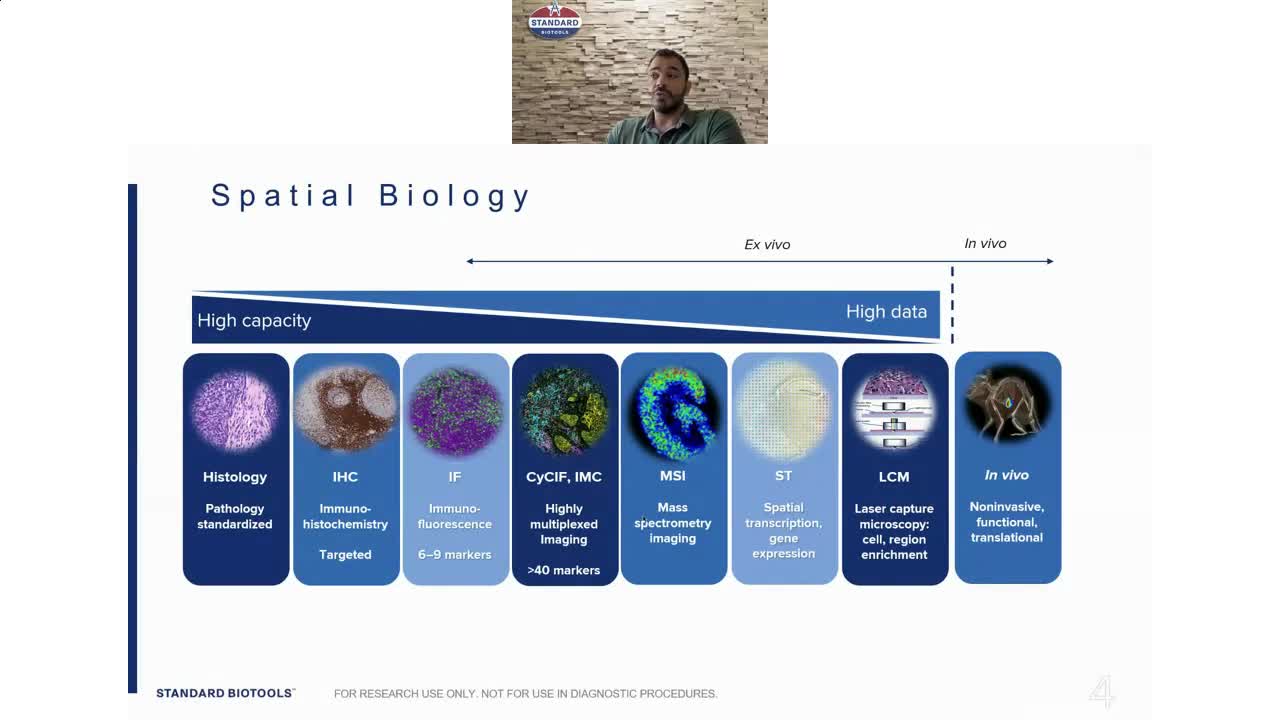
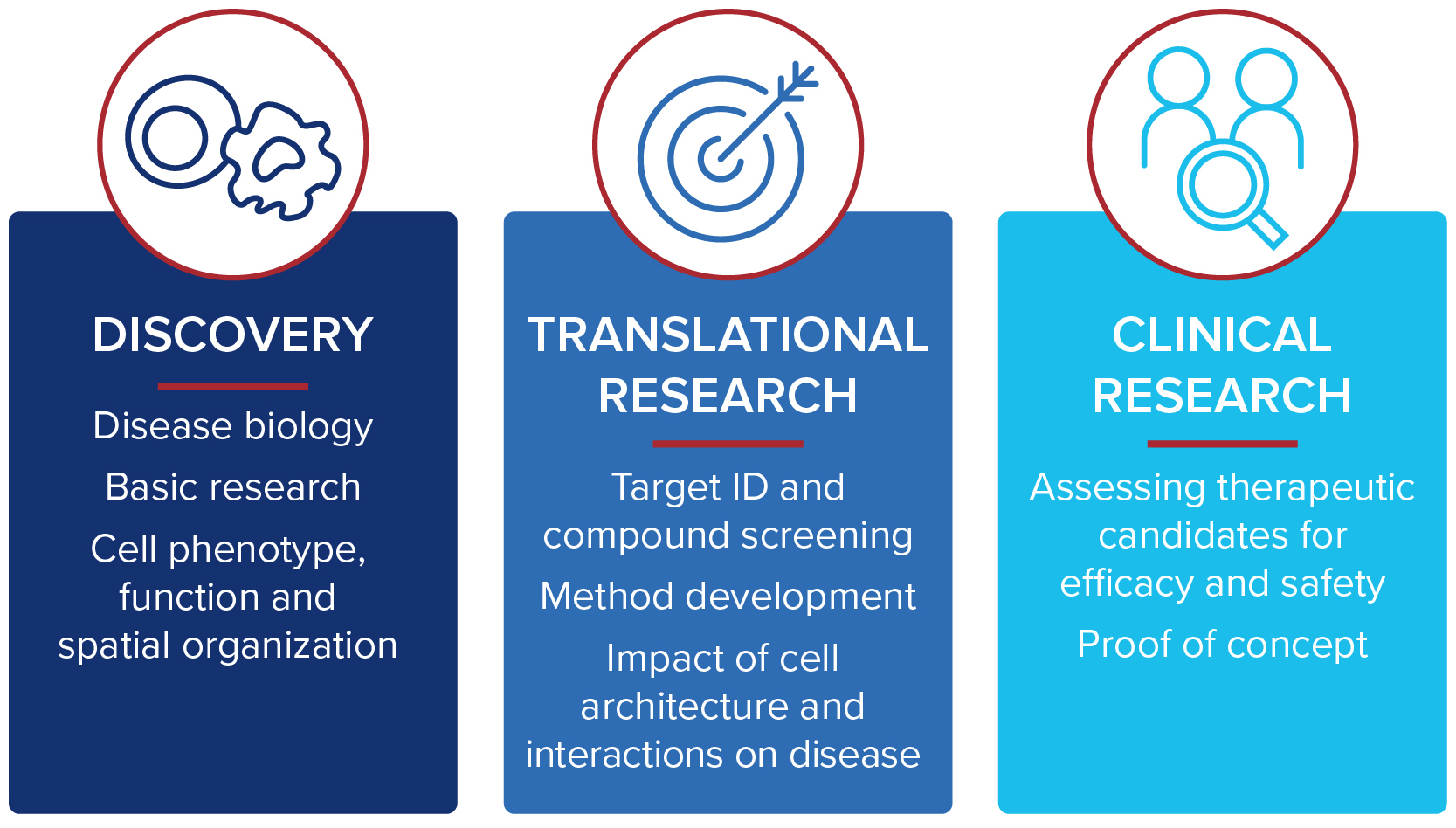
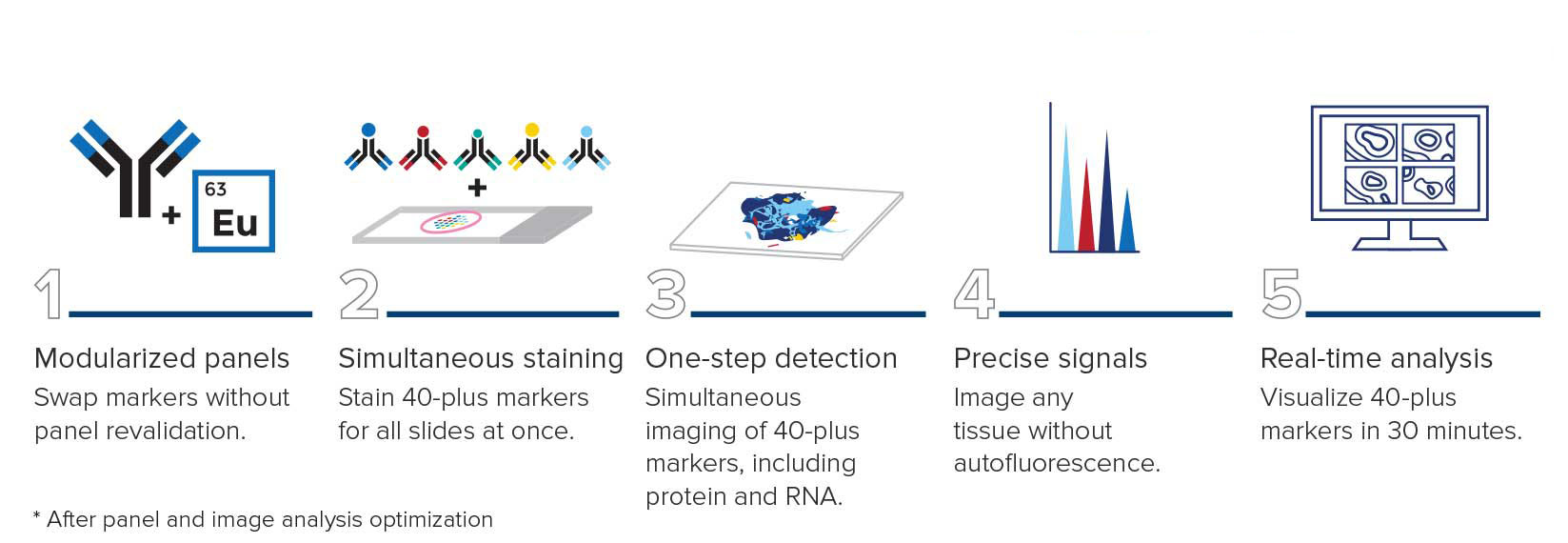
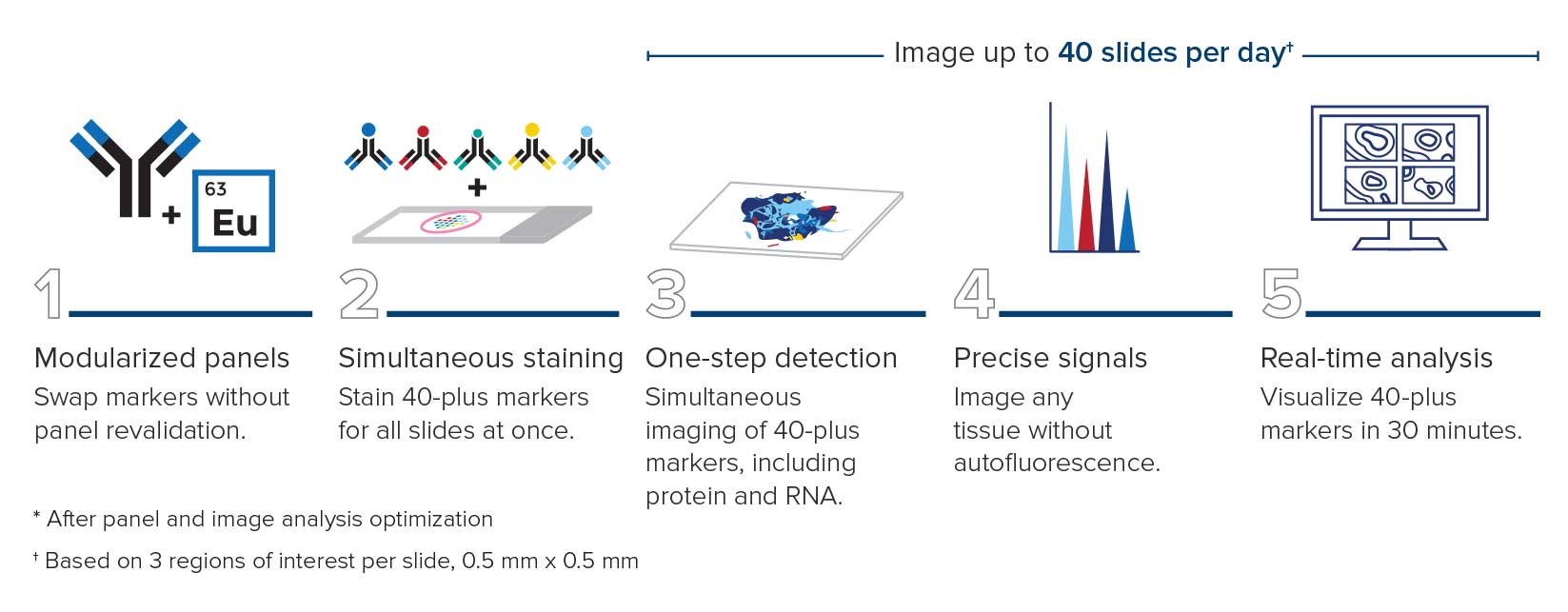
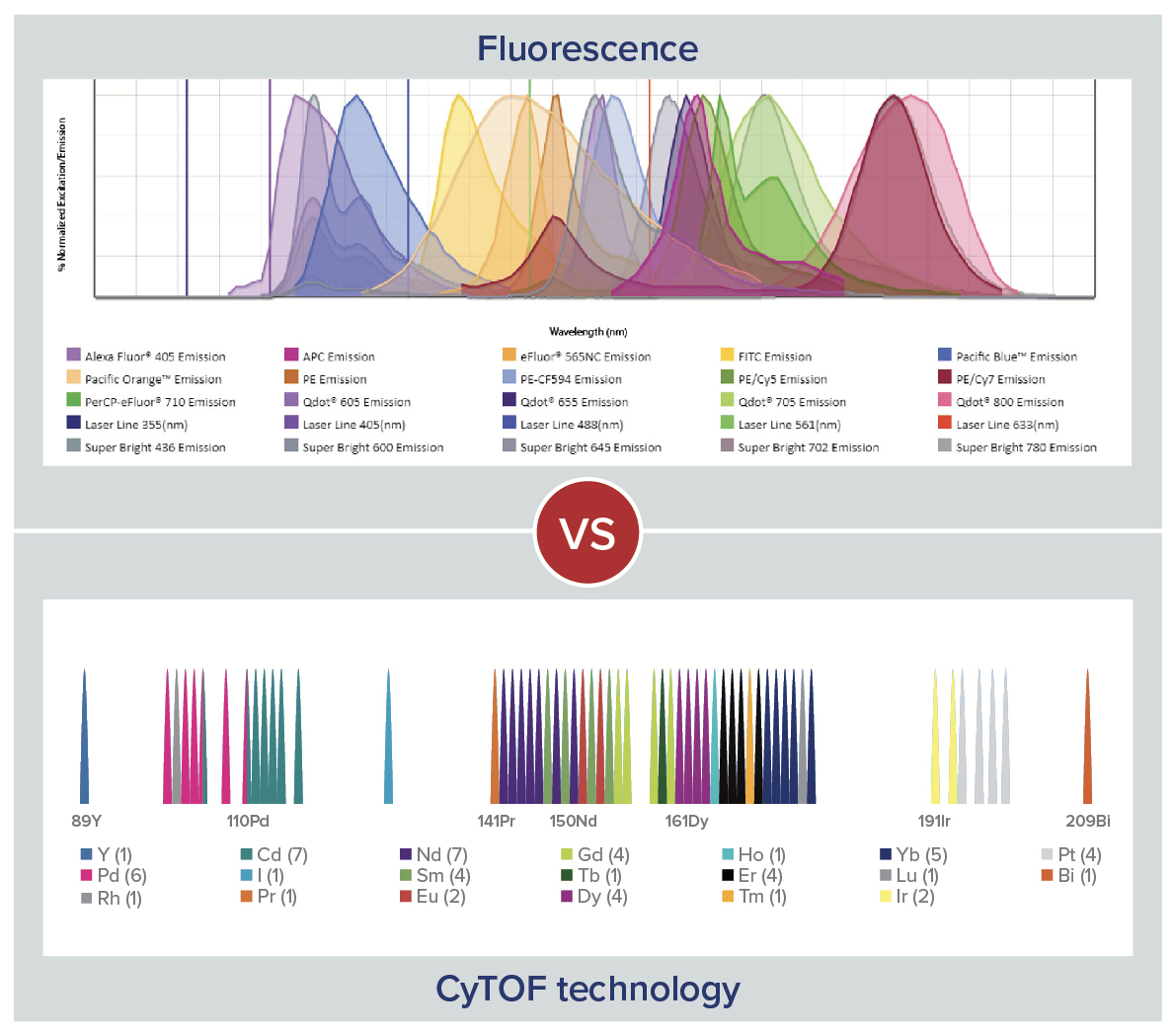
Traditional pathology methods such as immunohistochemistry are the gold standard in characterizing tissue morphology and confirming target molecule expression. When combined with genetic, RNA transcript and protein information, they can be used to decipher the molecular pathways that correlate to an observed phenotype.