We are pleased to announce that SomaLogic has merged with Standard BioTools™.
With this new broader organization comes a comprehensive suite of complementary high-integrity solutions to support your research, allowing us to serve you and your needs in an even greater capacity with access to genomics, proteomics and imaging tools.

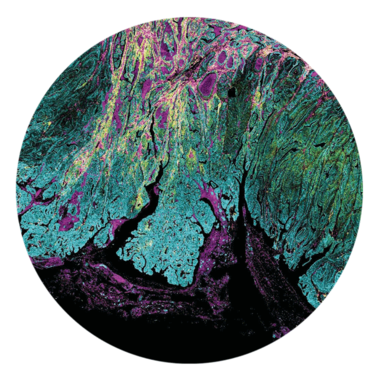
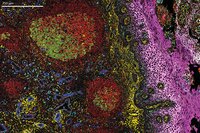
Hyperion XTi Imaging System
Unlocking New Views in Spatial Biology
Introducing new groundbreaking capabilities for multiplexed tissue imaging at never-before-seen speeds and automation. Discover the throughput and precision that is revolutionizing spatial biology.
Learn more
Apr 25 - Apr 26
Location
Immuno 2024 in London, UK
May 03
Location
CyTOF Summit | Flow into the Future: Cytometry and Spatial Biology Explored in Edinburgh, Scotland
May 03 - May 07
Location
Immunology 2024 (American Association of Immunologists) in Chicago, IL
May 04 - May 08
Location
CYTO (ISAC International Society for the Advancement of Cytometry) in Edinburgh, Scotland
May 06 - May 09
Location
APHL (Association of Public Health Laboratories) in Milwaukee, WI
May 06 - May 10
Location
WRIB (Workshop on Recent Issues in Bioanalysis) in San Antonio, TX
May 08 - May 10
Location
Precision Med Exhibition and Summit International in Dubai, UAE
May 14 - May 17
Location
EBW Congress (Europe Biobank Week) in Vienna, Austria
May 19
Location
UCSF BayViro Symposium (Bay Area Virus Network) in Berkeley, CA
May 30 - May 31
Location
VIB | Recent Insights into Immuno-Oncology (2nd Edition) in Antwerpen, Belgium

CUSTOMER STORIES
Our customers rely on our groundbreaking technology to transform their insights into actionable data.
SEE THE RESULTSNEW FEATURED INSTRUMENT
Biomark X9™ System for High-Throughput Genomics
The only genomics system for real-time PCR and NGS library preparation to support discovery through screening.
Deep Insights With Nanoscale Genomics


REIMAGINE CYTOMETRY
SEE THE NEXT EVOLUTIONDISCOVER THE POWER OF CRYSTAL-CLEAR INSIGHTS
Unless explicitly and expressly stated otherwise, all products are provided for Research Use Only, not for use in diagnostic procedures. Find more information here.